Deep sequencing to study genome replication
Recent advances in DNA sequencing technology are revolutionising modern biology. We are using these technologies to study the order in which genomes are replicated. When a DNA sequence replicates one copy gives rise to two copies and this change in DNA copy number can be directly measured by the new sequencing technologies - referred to as 'deep sequencing'. Using such methods we are able to precisely measure when each part of the genome replicates; from those regions that replicate first to those that are latest. The order in which a genome is replicated is normally tightly regulated and we have found that changing the time at which regions replicate can have pathological consequences (Natsume et al., 2013).
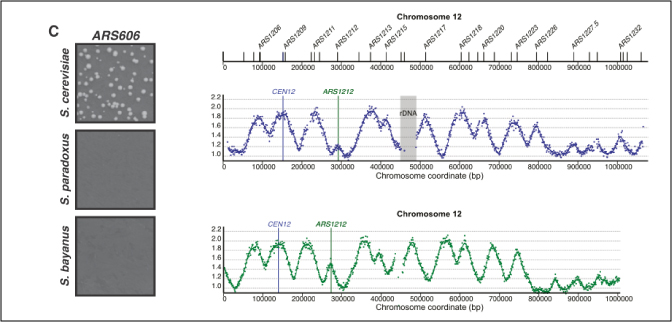
We have developed a complementary deep sequencing approaches to measure the dynamics of genome replication in a range of organisms. These methods measure the increase in DNA copy number as a sequence replicates. In eukaryotes, replicating cells generally need to be enriched and we have achieved this using fluorescence activated cell sorting (FACS) or cell cycle synchronisation. By contrast, in prokaryotes we have been able to measure replication dynamics directly in exponentially growing cultures due to the high proportion of cells actively engaged in genome replication. Using these approaches we have been able to learn about the basic cellular mechanisms that regulate genome replication. For example, in S. cerevisiae we have found that the centromere promotes the activity of proximal replication origins to ensure early centromere replication (Natsume et al., 2013). In E. coli we have shown that replication termination can give rise to new rounds of replication initiation (Rudolph et al., 2013).
Related publications
The dynamics of genome replication using deep sequencing.
Müller et al. (2014)
Nucleic Acids Res., 42(1):e3
Avoiding chromosome pathology when replication forks collide.
Rudolph et al. (2013)
Nature, 500(7464):608-11
Kinetochores coordinate pericentromeric cohesion and early DNA replication by cdc7-dbf4 kinase recruitment.
Natsume et al. (2013)
Mol. Cell, 50(5):661-74
High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome.
Liti et al. (2013)
BMC Genomics, 14():69
Conservation of replication timing reveals global and local regulation of replication origin activity.
Müller & Nieduszynski (2012)
Genome Res., 22(10):1953-62