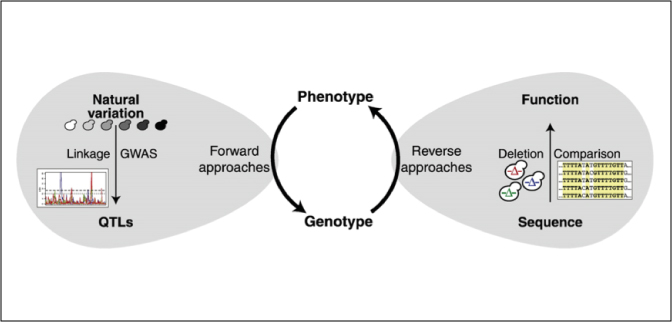
From sequence to function: Insights from natural variation in budding yeasts.
Conrad A Nieduszynski, Gianni Liti
Biochim. Biophys. Acta (2011), 1810(10):959-66![]() | PubMed | PubMed Central | Biochim. Biophys. Acta
| PubMed | PubMed Central | Biochim. Biophys. Acta
Natural variation offers a powerful approach for assigning function to DNA sequence-a pressing challenge in the age of high throughput sequencing technologies.
Our publications
Click to expand
2017
DNA replication timing influences gene expression level.
Müller & Nieduszynski (2017)
J. Cell Biol., 216(7):1907-1914
Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome.
Shen et al. (2017)
Science, 355(6329):
2015
Discovery of an Unconventional Centromere in Budding Yeast Redefines Evolution of Point Centromeres.
Kobayashi et al. (2015)
Curr. Biol., 25(15):2026-33
A global profile of replicative polymerase usage.
Daigaku et al. (2015)
Nat. Struct. Mol. Biol., 22(3):192-8
2014
The dynamics of genome replication using deep sequencing.
Müller et al. (2014)
Nucleic Acids Res., 42(1):e3
2013
High-resolution replication profiles define the stochastic nature of genome replication initiation and termination.
Hawkins et al. (2013)
Cell Rep, 5(4):1132-41
Accelerated growth in the absence of DNA replication origins.
Hawkins et al. (2013)
Nature, 503(7477):544-547
Stochastic association of neighboring replicons creates replication factories in budding yeast.
Saner et al. (2013)
J. Cell Biol., 202(7):1001-1012
A Link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast.
Hoggard et al. (2013)
PLoS Genet., 9(9):e1003798
Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts.
Newman et al. (2013)
Nucleic Acids Res., 41(21):9705-18
Avoiding chromosome pathology when replication forks collide.
Rudolph et al. (2013)
Nature, 500(7464):608-11
Kinetochores coordinate pericentromeric cohesion and early DNA replication by cdc7-dbf4 kinase recruitment.
Natsume et al. (2013)
Mol. Cell, 50(5):661-74
High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome.
Liti et al. (2013)
BMC Genomics, 14():69
2012
Mathematical modeling of genome replication.
Retkute et al. (2012)
Phys Rev E Stat Nonlin Soft Matter Phys, 86(3 Pt 1):031916
Conservation of replication timing reveals global and local regulation of replication origin activity.
Müller & Nieduszynski (2012)
Genome Res., 22(10):1953-62
A putative homologue of CDC20/CDH1 in the malaria parasite is essential for male gamete development.
Guttery et al. (2012)
PLoS Pathog., 8(2):e1002554
2011
OriDB, the DNA replication origin database updated and extended.
Siow et al. (2011)
Nucleic Acids Res., 40(Database issue):D682-6
Dynamics of DNA replication in yeast.
Retkute et al. (2011)
Phys. Rev. Lett., 107(6):068103
Comparative functional genomics of the fission yeasts.
Rhind et al. (2011)
Science, 332(6032):930-6
From sequence to function: Insights from natural variation in budding yeasts.
Nieduszynski & Liti (2011)
Biochim. Biophys. Acta, 1810(10):959-66
2010
Mathematical modelling of whole chromosome replication.
de Moura et al. (2010)
Nucleic Acids Res., 38(17):5623-33
2009
The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication.
Shor et al. (2009)
PLoS Genet., 5(12):e1000755
Detection of replication origins using comparative genomics and recombinational ARS assay.
Nieduszynski & Donaldson (2009)
Methods Mol. Biol., 521():295-313
2008
Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site.
Chang et al. (2008)
Mol. Cell. Biol., 28(16):5071-81
2006
OriDB: a DNA replication origin database.
Nieduszynski et al. (2006)
Nucleic Acids Res., 35(Database issue):D40-6
Genome-wide identification of replication origins in yeast by comparative genomics.
Nieduszynski et al. (2006)
Genes Dev., 20(14):1874-9
2005
The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription.
Nieduszynski et al. (2005)
Nucleic Acids Res., 33(8):2410-20
2004
The cyclin A1-CDK2 complex regulates DNA double-strand break repair.
Müller-Tidow et al. (2004)
Mol. Cell. Biol., 24(20):8917-28
Cyclin A1 protein shows haplo-insufficiency for normal fertility in male mice.
van der Meer et al. (2004)
Reproduction, 127(4):503-11
2002
Whole-genome analysis of animal A- and B-type cyclins.
Nieduszynski et al. (2002)
Genome Biol., 3(12):RESEARCH0070
Ku complex controls the replication time of DNA in telomere regions.
Cosgrove et al. (2002)
Genes Dev., 16(19):2485-90