Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts.
Timothy J Newman, Mohammed A Mamun, Conrad A Nieduszynski, J Julian Blow
Nucleic Acids Res. (2013), 41(21):9705-18![]() | PubMed | PubMed Central | Nucleic Acids Res.
| PubMed | PubMed Central | Nucleic Acids Res.
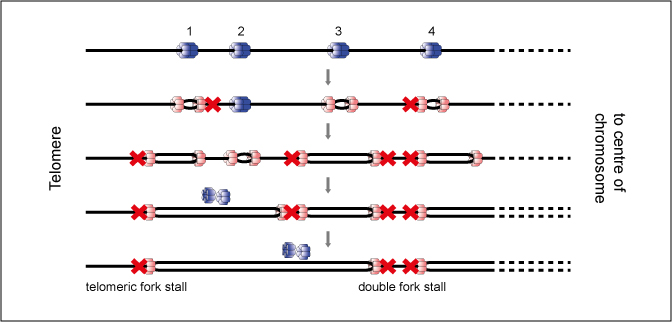
During S phase, the entire genome must be precisely duplicated, with no sections of DNA left unreplicated. Here, we develop a simple mathematical model to describe the probability of replication failing due to the irreversible stalling of replication forks. We show that the probability of complete genome replication is maximized if replication origins are evenly spaced, the largest inter-origin distances are minimized, and the end-most origins are positioned close to chromosome ends. We show that origin positions in the yeast Saccharomyces cerevisiae genome conform to all three predictions thereby maximizing the probability of complete replication if replication forks stall. Origin positions in four other yeasts-Kluyveromyces lactis, Lachancea kluyveri, Lachancea waltii and Schizosaccharomyces pombe-also conform to these predictions. Equating failure rates at chromosome ends with those in chromosome interiors gives a mean per nucleotide fork stall rate of ∼5 × 10(-8), which is consistent with experimental estimates. Using this value in our theoretical predictions gives replication failure rates that are consistent with data from replication origin knockout experiments. Our theory also predicts that significantly larger genomes, such as those of mammals, will experience a much greater probability of replication failure genome-wide, and therefore will likely require additional compensatory mechanisms.
Our publications
Click to expand
2017
DNA replication timing influences gene expression level.
Müller & Nieduszynski (2017)
J. Cell Biol., 216(7):1907-1914
Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome.
Shen et al. (2017)
Science, 355(6329):
2015
Discovery of an Unconventional Centromere in Budding Yeast Redefines Evolution of Point Centromeres.
Kobayashi et al. (2015)
Curr. Biol., 25(15):2026-33
A global profile of replicative polymerase usage.
Daigaku et al. (2015)
Nat. Struct. Mol. Biol., 22(3):192-8
2014
The dynamics of genome replication using deep sequencing.
Müller et al. (2014)
Nucleic Acids Res., 42(1):e3
2013
High-resolution replication profiles define the stochastic nature of genome replication initiation and termination.
Hawkins et al. (2013)
Cell Rep, 5(4):1132-41
Accelerated growth in the absence of DNA replication origins.
Hawkins et al. (2013)
Nature, 503(7477):544-547
Stochastic association of neighboring replicons creates replication factories in budding yeast.
Saner et al. (2013)
J. Cell Biol., 202(7):1001-1012
A Link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast.
Hoggard et al. (2013)
PLoS Genet., 9(9):e1003798
Replisome stall events have shaped the distribution of replication origins in the genomes of yeasts.
Newman et al. (2013)
Nucleic Acids Res., 41(21):9705-18
Avoiding chromosome pathology when replication forks collide.
Rudolph et al. (2013)
Nature, 500(7464):608-11
Kinetochores coordinate pericentromeric cohesion and early DNA replication by cdc7-dbf4 kinase recruitment.
Natsume et al. (2013)
Mol. Cell, 50(5):661-74
High quality de novo sequencing and assembly of the Saccharomyces arboricolus genome.
Liti et al. (2013)
BMC Genomics, 14():69
2012
Mathematical modeling of genome replication.
Retkute et al. (2012)
Phys Rev E Stat Nonlin Soft Matter Phys, 86(3 Pt 1):031916
Conservation of replication timing reveals global and local regulation of replication origin activity.
Müller & Nieduszynski (2012)
Genome Res., 22(10):1953-62
A putative homologue of CDC20/CDH1 in the malaria parasite is essential for male gamete development.
Guttery et al. (2012)
PLoS Pathog., 8(2):e1002554
2011
OriDB, the DNA replication origin database updated and extended.
Siow et al. (2011)
Nucleic Acids Res., 40(Database issue):D682-6
Dynamics of DNA replication in yeast.
Retkute et al. (2011)
Phys. Rev. Lett., 107(6):068103
Comparative functional genomics of the fission yeasts.
Rhind et al. (2011)
Science, 332(6032):930-6
From sequence to function: Insights from natural variation in budding yeasts.
Nieduszynski & Liti (2011)
Biochim. Biophys. Acta, 1810(10):959-66
2010
Mathematical modelling of whole chromosome replication.
de Moura et al. (2010)
Nucleic Acids Res., 38(17):5623-33
2009
The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication.
Shor et al. (2009)
PLoS Genet., 5(12):e1000755
Detection of replication origins using comparative genomics and recombinational ARS assay.
Nieduszynski & Donaldson (2009)
Methods Mol. Biol., 521():295-313
2008
Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site.
Chang et al. (2008)
Mol. Cell. Biol., 28(16):5071-81
2006
OriDB: a DNA replication origin database.
Nieduszynski et al. (2006)
Nucleic Acids Res., 35(Database issue):D40-6
Genome-wide identification of replication origins in yeast by comparative genomics.
Nieduszynski et al. (2006)
Genes Dev., 20(14):1874-9
2005
The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription.
Nieduszynski et al. (2005)
Nucleic Acids Res., 33(8):2410-20
2004
The cyclin A1-CDK2 complex regulates DNA double-strand break repair.
Müller-Tidow et al. (2004)
Mol. Cell. Biol., 24(20):8917-28
Cyclin A1 protein shows haplo-insufficiency for normal fertility in male mice.
van der Meer et al. (2004)
Reproduction, 127(4):503-11
2002
Whole-genome analysis of animal A- and B-type cyclins.
Nieduszynski et al. (2002)
Genome Biol., 3(12):RESEARCH0070
Ku complex controls the replication time of DNA in telomere regions.
Cosgrove et al. (2002)
Genes Dev., 16(19):2485-90